No cell exists in isolation. Every cell has a context. Cells are surrounded by nutrients, extracellular matrix, and crucially: other cells.
I’m interested in how cells communicate with each other. Unfortunately, empirical analysis of cell-cell communication in a multicellular environment is limited. This is largely due to the technical difficulty in resolving between different cell lineages in a co-culture.
Here’s the problem: Proteins from one cell type are indistinguishable to those from a different cell type. Proteins from two different cells may have different origins – and therefore contain different information – but they are biochemically identical. This complicates the investigation of multicellular signalling because we have no spacial resolution of a spacial system.
For example, let's say we're interested in how cells in breast cancer communicate with each other. I have epithelial cancer cells and mammary fibroblasts. I mix the epithelial cells with the fibroblasts and leave them to exchange signals. I lyse the co-culture, perform a traditional western blot for my favourite signalling node and get a lovely bright band. Unfortunately, I can’t tell if the band came from the cancer cells or the fibroblasts. My western blot antibody can not resolve between the same signalling node from the two different cells types. I have no spacial contrast.
What we need is a way to resolve between two different cell types. We need cell-specific data.
Last year Gauthier et al (Nature Methods, 2013) reported a potential solution to this problem. The authors called it “Cell Type–specific labelling using Amino acid Precursors” (or CTAP). It’s very clever.
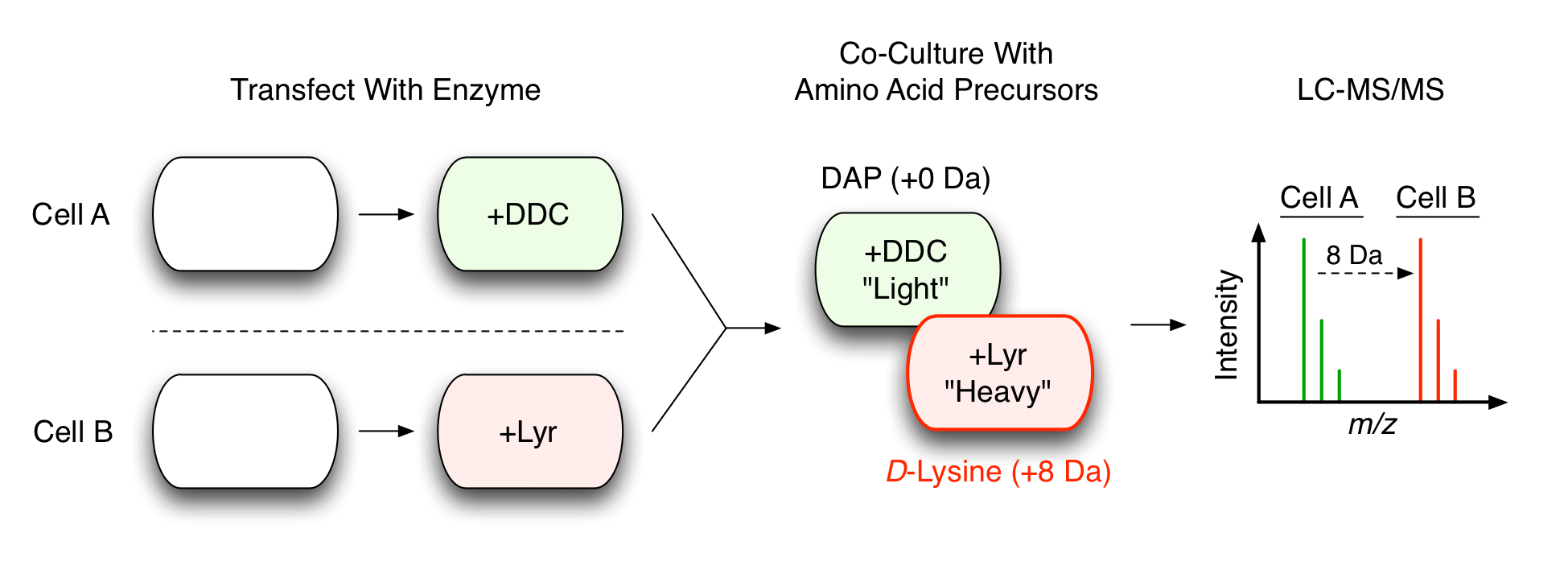
CTAP uses non-mammalian amino acid processing enzymes to convert lysine-precursors into L-lysine. When expressed in mammalian cells, these enzymes convert their lysine-precursors into into L-lysine and the newly formed L-lysine is incorporated into every protein in the cell.
Now here’s the first clever bit: There are two enzymes.
One enzyme (Lyr) converts a specific precursor (called D-lysine) into L-lysine. The other enzyme (DDC) converts a different precursor (called DAP) into L-lysine. Thus, we have a pair of enzymes capable of producing the same product from two different substrates.
The second clever bit: The amino acid precursors can be labeled.
D-lysine can be isotopically labeled “heavy”. The Lyr enzyme can then convert “heavy” D-lysine into “heavy” L-lysine. DAP (the other precursor) can remain unlabeled – or “light”. As a result, the L-lysine produced from DDC is “light” and the L-lysine produced by Lyr is “heavy”. Both enzymes produce L-lysine but now we have isotopic resolution between the two outputs. We have contrast.
Let’s return to our breast cancer experiment.
This time, we express the DDC enzyme in our breast cancer cells and the Lyr enzyme in our mammary fibroblasts. The cells are then mixed together and grown in the presence of both “light” DAP and “heavy” D-lysine. In the breast cancer cells, our DDC enzyme converts “light” DAP into “light” L-lysine. As the breast cancer cells don’t have the Lyr enzyme, they can’t use the “heavy” D-lysine. As a result, all the proteins in the breast cancer cells are now “light”. Over in the fibroblasts however, Lyr is converting “heavy” D-lysine into "heavy" L-lysine. The fibroblasts don’t have the DDC enzyme so they can’t use the “light” DAP. Consequently, all the proteins in the fibroblasts are now “heavy”. We have “light” cancer cells and “heavy” fibroblasts.
This approach is conceptually similar to SILAC. The major difference is that whilst SILAC can be used to distinguish different cell types in transient co-cultures (see Jorgensen et al., 2009), CTAP permits the analysis of long-term, continuous co-cultures.
Now we leave our cells to communicate. But this time – instead of running a western blot to investigate our favourite signaling node – this time, we use mass-spectrometry. The mass-spectrometer can easily resolve between the “light” and “heavy” proteins. As a result, we can easily distinguish between cancer cell proteins (they’re all “light”) and fibroblast proteins (they’re all “heavy”). We have cell-specific data. We have contrast.
This concept is illustrated below:
CTAP uses cell-specific amino acid processing enzymes (DDC and Lyr) to isotopically label cells. When the co-culture is analysed by mass-sepctrometry (LC-MS/MS), we can easily distinguish between the proteins from each cell.
It’s a great idea.
I work on cell-cell communication in Pancreatic Ductal Adenocarcinoma (PDAC) and when I first read about CTAP it really excited me. As a protein biochemist I loved the idea of using enzymes to confer cell-specific labeling and at the ICR I’m privileged to have access to a mass-spectrometer capable of analysing CTAP experiments.
So last year I decided to try CTAP for myself. I ordered some enzymes and transfected them into my cells. Unfortunately, at first, I couldn’t really get it to work. My DDC enzyme didn’t confer DAP-dependent proliferation and my Lyr enzyme kept flying out of the cells. In theory it should have worked – but the enzymes were letting me down.
CTAP could really change the way I investigate PDAC cell-cell communication so this was extremely frustrating. I knew the idea of CTAP was great but the pragmatic reality was disappointing.
So I decided to come-up with a new pair of CTAP enzymes. I screened a panel of alternative enzymes and engineered the best ones to work more efficiently for CTAP. With help from some colleagues in the Jorgensen Lab, I tested these enzymes and found them to be excellent for CTAP. We ended up with a lovely pair of new CTAP enzymes.
This little endeavor has just been published as an open access article in Molecular and Cellular Proteomics under the title: “Cell-Specific Labeling Enzymes For Analysis of Cell-Cell Communication in Continuous Co-Culture”. I've deposited the enzyme pair in AddGene so anyone interested in CTAP can use them. If you're interested in cell-specific labelling, take a look.